Edit this page
Open an issue
Data Import
Warning
If you have not installed trajectoryGuide into 3D Slicer please follow the installation instructions.
The first module in trajectoryGuide handles the import of patient imaging data. The data should be within a single directory, this directory will be selected within the import window (do not select the files within the directory). During the initial data import for a patient, trajectoryGuide will store a copy of the imaging data into a source directory as a backup, these files will remain unchanged.
trajectoryGuide stores all imaging data in NIFTI (Neuroinformatics Technology Initiative) format, with the file extension .nii.gz. If the original imaging data is in DICOM format, the files will first need to be converted to NIFTI according to BIDS (see section above).
Data directory¶
Input directory structure¶
trajectoryGuide requires the input data folder to be organized according to Brain Imaging Data Structure (BIDS). The following is an example input directory.
bids/
├── dataset_description.json
└── sub-<subject_label>/
└── ses-<ses_label>/
├── anat/
│ ├── sub-<subject_label>_ses-<ses_label>_T1w.nii.gz
│ ├── sub-<subject_label>_ses-<ses_label>_PD.nii.gz
│ └── sub-<subject_label>_ses-<ses_label>_acq-Tra_T2w.nii.gz
└── ct/
└── sub-<subject_label>_ses-<ses_label>_acq-Frame_ct.nii.gz
Output directory structure¶
trajectoryGuide trajectoryGuide stores processed data within the derivatives directoy of the BIDS dataset.
bids/
├── dataset_description.json
└── sub-<subject_label>/...
derivatives/
└── trajectoryGuide/
└── sub-<subject_label>/
├── sub-<subject_label>_surgical_data.json
├── sub-<subject_label>_T1w.nii.gz
├── sub-<subject_label>_T1w.json
├── sub-<subject_label>_space-T1w_acq-Tra_T2w.nii.gz
├── sub-<subject_label>_space-T1w_acq-Tra_T2w.json
├── sub-<subject_label>_desc-rigid_from-TraT2w_to-T1w_xfm.h5
├── sub-<subject_label>_space-T1w_PD.nii.gz
├── sub-<subject_label>_space-T1w_PD.json
├── sub-<subject_label>_desc-rigid_from-PD_to-T1w_xfm.h5
├── sub-<subject_label>_ses-pre_coordsystem.json
├── settings/...
├── source/...
├── space/...
└── summaries/...
Each image volume has an associated .json file that stores metadata associated with the volume as it progresses through the trajectoryGuide workflow. Select the import options prior to loading the patient directory.
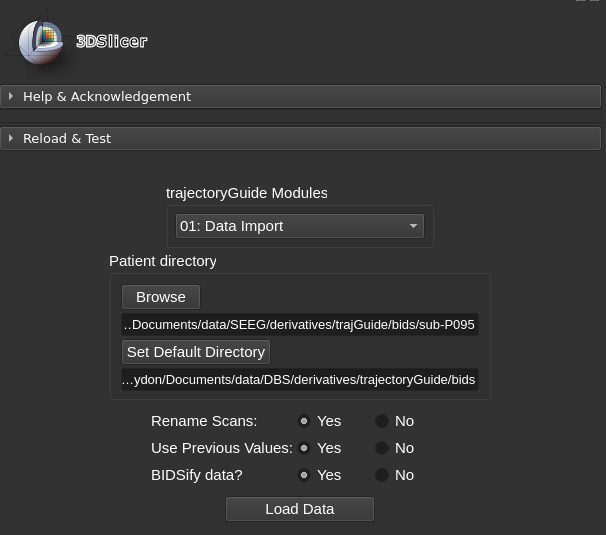
- Rename Scans: trajectoryGuide will rename the imaging data to comply with BIDS format, the imaging filenames will be shortened
- Use Previous Values: this option is recommended. If the patient directory has already been loaded by trajectoryGuide, then the previous data values will be re-loaded
- BIDSify data: future feature to convert DICOM data to BIDS.